News //
This feature is written by Everlyn Julya Koh, who undertook an internship with the butterfly digitisation project.

In 2019, the museum embarked on a butterfly digitisation project with an aim to create a virtual collection of butterflies from Singapore and Peninsular Malaysia, thanks to the co-funding from Biodiversity Information Fund for Asia (BIFA) by the Ministry of Environment, Government of Japan and the museum. After more than a year of hard work, we are proud to share the data of 12,702 butterfly specimens from our entomological collection with the Global Biodiversity Information Facility (GBIF), an open-access international biodiversity database.
We digitised 12,702 specimens, comprising 978 species and 1,112 subspecies from six families. These specimens were collected across 187 known localities in Singapore and Peninsular Malaysia over 63 years (1936–1998). Among the butterflies digitised, 319 species are considered to be rare or very rare.
Specimens and their labels carry a wealth of scientific information in the form of taxonomic data (e.g. species name), occurrence data (e.g. place of collection) and temporal data (e.g. date of collection). Collectively, these historical data form a baseline measure of biodiversity during a given period of time from which researchers can note changes by comparing with recent observations.

Most butterflies digitised in this project were collected from 1960 to 1990, a period in which Singapore and Peninsular Malaysia experienced extensive landscape changes due to human development. By sharing specimen data from this period globally, we hope to encourage scientific studies into the extraordinary butterfly diversity of this region. The data can prove valuable in analysing the impacts of environmental modifications on the butterfly diversity in this region, and in bridging the historical and current knowledge of butterfly species here.
The butterfly specimen digitisation process converts information associated with physical butterfly specimens into a digital record—a format which is more conducive for online sharing. Such a specimen digitisation workflow typically involves the transcription of label information into datasets, specimen imaging (i.e. macro photography of specimen) and the georeferencing of locality information (i.e. adding geographical data to an image such as a map).
With the sheer number of specimens involved, the specimen digitisation process had been a rather arduous one. Despite that, we still found great joy in learning about butterflies through the meticulous handling of specimens and in the proper care of dry specimens.

In this blog post, we highlight three interesting aspects of the butterfly digitisation process that left a deep impression on us.
In warm and humid tropical countries like Singapore and Malaysia, mould can grow on specimens that are not kept in a cool and dry environment. This is problematic as the fungi can grow deeply into and feed extensively on the infected specimen, degrading it with time. For this project, it is definitely not ideal to photograph specimens that have their key features covered in a thick layer of mould!
Prior to specimen imaging, we inspected specimens for mould growth and thoroughly cleaned any mouldy specimens. Fortunately, only a handful of specimens required cleaning.

It can take up to 30 minutes to clean a specimen. We gently brush mouldy areas with a small amount of alcohol to remove and kill existing mould and spores. As mouldy specimens are especially fragile, a great deal of focus is required to prevent damage.
Look at the stark difference specimen cleaning can make! Not only will proper specimen maintenance and preservation techniques contribute towards specimens having a longer shelf-life, the specimens now have their defining features clearly visible for imaging at a later stage of our project!
Of all the steps involved in the digitisation workflow, the team spent the most time on specimen imaging. This is warranted as it is important that specimen images taken are of a high quality (i.e. well-exposed and focused images with key identifying features of the specimen clearly shown) since they can potentially serve as powerful visual references for species identification.
In this project, specimen imaging was most efficient as pairwork. As depicted in the image below, one member photographs a specimen (left) while the other member returns a photographed specimen into the specimen drawer and prepares the next specimen to be photographed in an imaging tray (right).

In an imaging tray, we suspended the relatively flat butterfly specimen on the same height with all its labels using nylon strings. This arrangement ensures that all elements in the imaging tray are in the same depth of field and thus eliminates the need for individual focus adjustments. A pair of forceps was used when placing original specimen labels into the imaging tray so as to prevent the oil from our skin from damaging it.

At the imaging station, we photographed both the dorsal and ventral sides of each specimen, and made sure that the specimen and its labels are well-aligned and in focus.


All in all, with each specimen needing at least three minutes to be imaged, it would have taken the team at least 635 hours (26.5 continuous days) to photograph all 12,702 specimens in one sitting!
With the massive amount of data and images generated from this project, applying changes to thousands of files manually was a time-consuming and painful task. Automating certain key processes in the specimen digitisation process was definitely a game changer!
A significant use of automation for this project was in the batch-renaming of image files. When an image is taken, the camera saves the image file with an uninformative name (e.g. IMG_001) that tells us nothing about what is in the photograph. Ultimately, all image file names need to be changed to state the identity of the specimen photographed, as well as the side of the specimen photographed (i.e. dorsal or ventral). Once renamed, we can then easily check for missing or duplicated images at a glance without having to open and view the images individually.

It would have taken the team at least 10 seconds to manually rename each image file. This can become a considerable burden with the sheer number of image files to be renamed in this project!
Thus, for all raw images, we ran an algorithm that uses Optical Character Recognition (OCR) to first scan or “read” the raw images to identify: (1) the unique specimen identifier and (2) D/V labels. Once successfully identified, the algorithm automatically renames each image file according to what has been captured in the image (e.g. ZRC_ENT00026645-Dorsal.CR2).

This algorithm was consistently optimized throughout the project and was eventually able to recognise and rename the identifier code and D/V labels correctly 99% of the time. This meant that the team could spend more time getting more specimens imaged!
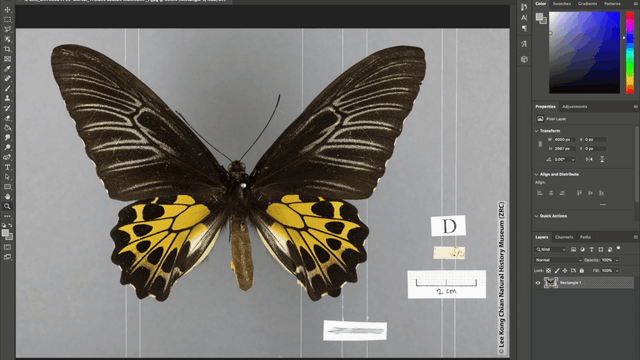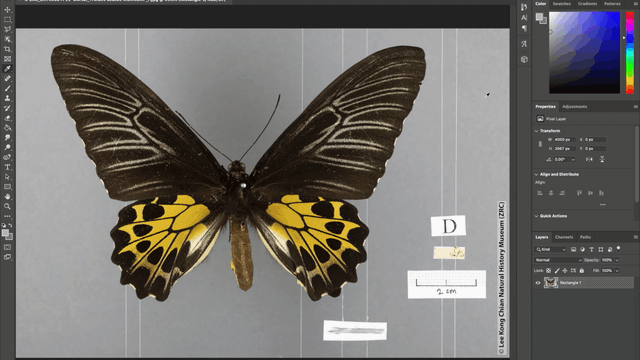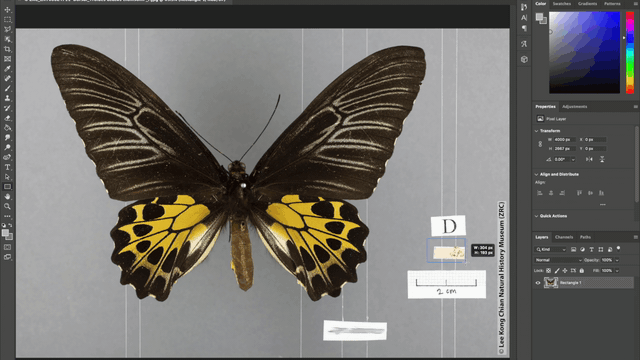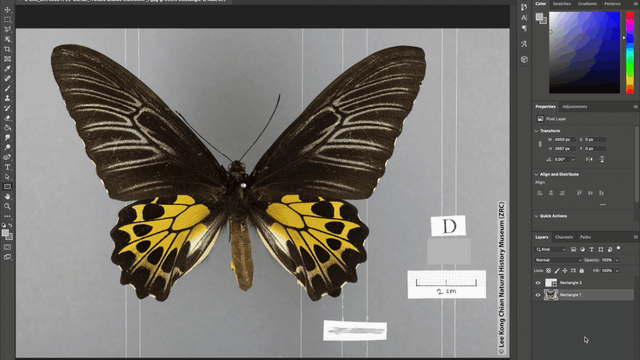
Additionally, the team also automated the photo-editing process such as the addition of watermarks to every image, as well as the redaction of locality labels in images of rare species. These automations were achieved using the “Action” tool in Adobe Photoshop where batches of image files can be automatically edited according to a fixed set of instructions applied. For example, in redacting locality information for images of rare specimens, a set of instructions to place a grey rectangle over the locality label was applied to all the images that need editing and voila, all the locality labels are covered up!
With these automations in the project, we saved close to 470 hours in total that was put to better use in improving the digital collection!
Our digitisation efforts culminated in ~24,000 specimen images and six datasets of butterflies from Singapore and Peninsular Malaysia which we proudly share with the world on the GBIF portal. Even with the automation of certain processes, the butterfly digitisation project has been a trying one with months after months of meticulous label transcription, specimen imaging and proof-reading of datasets. Looking back, it has definitely been a humbling learning experience and privilege for the team to be able to bring this project from inception to fruition.
In this digital age, the creation of online collections is a trajectory in which many museums in the world are taking today. Using current technology to accelerate and improve productivity in the museum, such a digitisation project will definitely not be the last of its kind in time to come. After all, an overarching goal of the museum is to modernize our collection and provide better access to our specimen data. Thus, by embracing new tools and technology, we hope that we can bring more from our museum’s collections to your home in the near future.
To access and make use of the digital butterfly data from this project, please visit Global Biodiversity Information Facility (GBIF) here. You can also check out the highlights of this project at the museum’s Instagram page here